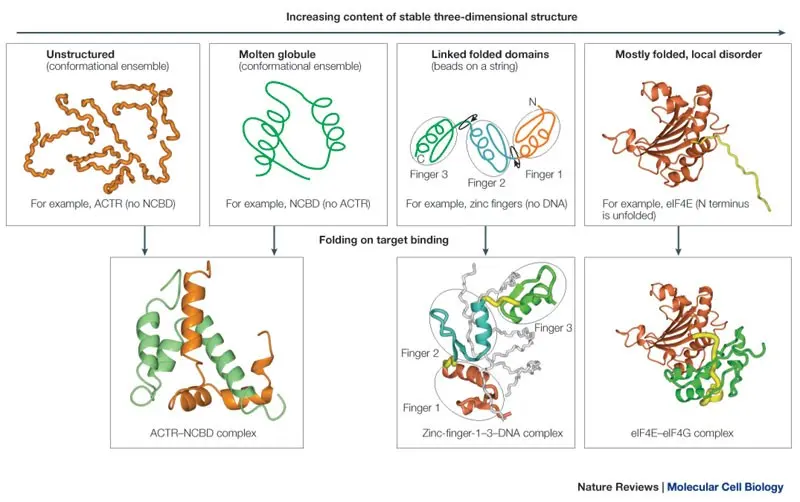
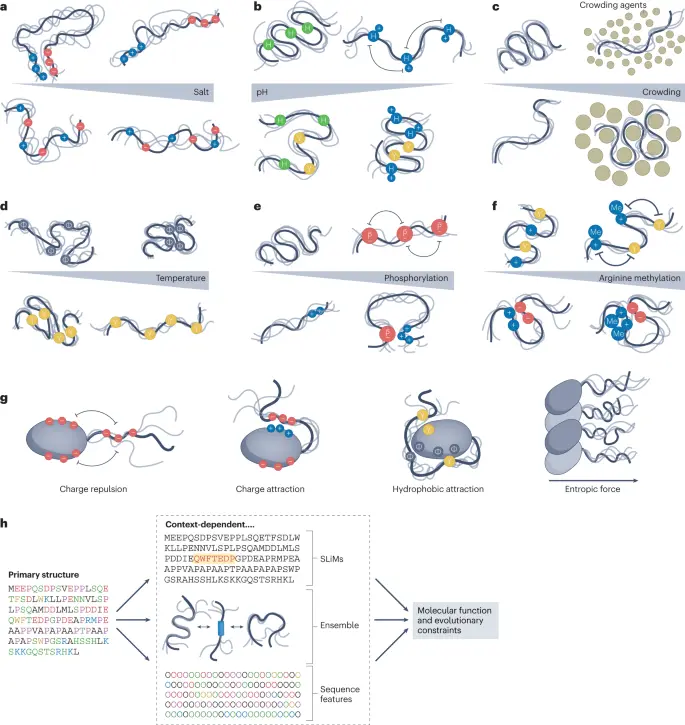
SLIRP: Structural Library of Intrinsic Residue Propensities
SLIRP is a key component of the Dynameomics database, developed by the University of Washington’s Department of Bioengineering. It provides detailed insights into the conformational behavior of amino acids under realistic simulation conditions.
What is SLIRP?
SLIRP is a dynamic library that compiles results from molecular dynamics (MD) simulations on GGXGG peptides. These peptides serve as models to study the intrinsic conformational preferences of amino acids, including alanine, glycine, serine, proline, and valine. Simulations also include protonated and deprotonated forms of acidic (Asp, Glu) and basic (His) amino acids, offering a comprehensive view of their dynamic behavior.
Objectives of SLIRP
Study Conformational Preferences: Analyze backbone torsion angles (phi, psi) to understand amino acid tendencies in protein environments.
Build Fragment Libraries: Develop simulation-based fragment libraries to improve protein structure modeling.
Analyze Flexibility: Identify dynamic regions of proteins using advanced flexibility analysis techniques.
SLIRP Database Content
The SLIRP database contains GGXGG peptide simulations at 298 K, representing the intrinsic conformational preferences of amino acids. Researchers can explore these data to enhance their understanding of protein dynamics and integrate it into structural modeling workflows.
Accessing SLIRP
SLIRP data is publicly accessible via the Dynameomics portal: http://www.dynameomics.org/shop. Users can explore simulations, download datasets, and apply the information to protein modeling and research.