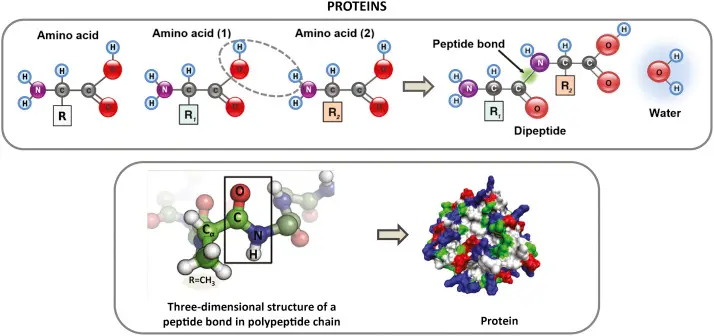
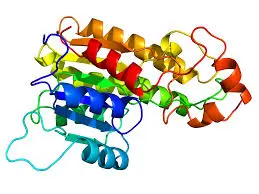
Molecular Dynamics Simulations of Proteins: Unlocking Protein Behavior with Dynameomics
Molecular dynamics simulations use physics-based models to calculate the forces and motions of atoms in a protein structure. By simulating these movements over time, scientists can:
Dynameomics:
Large-Scale MD Datasets
The Dynameomics database provides one of the largest collections of molecular dynamics simulations of proteins in the world. It contains:
Access high-quality MD datasets for comparative studies
Perform statistical analysis of protein behavior across many simulations
Integrate dynamic protein data into computational modeling and bioinformatics workflows
By leveraging Dynameomics data, scientists and bioengineers can:
Improve drug discovery pipelines by predicting protein-ligand interactions
Design stable enzymes for industrial and therapeutic applications
Study disease-related proteins to identify potential therapeutic targets